Deep generative model for Learning Streamline Embeddings from Tractography
This is a research project I’ve been working on since September 2022 in Dr. Paul Thompson’s lab at the Imaging Genetics Center at University of Southern California since my research rotation. Our goal is to utilize deep representation learning for analyzing tractography data for population analysis.
Method
Tractography generated from diffusion MRI are often used to study white matter pathways. Each whole brain tractogram can generate from 500K to 1M streamlines, making processing and downstream analyses very computationally expensive. In this work, we present a framework using deep representation learning based on a convolutional variational autoencoder (ConvVAE) to learn low-dimensional embeddings of white matter tracts.
Result
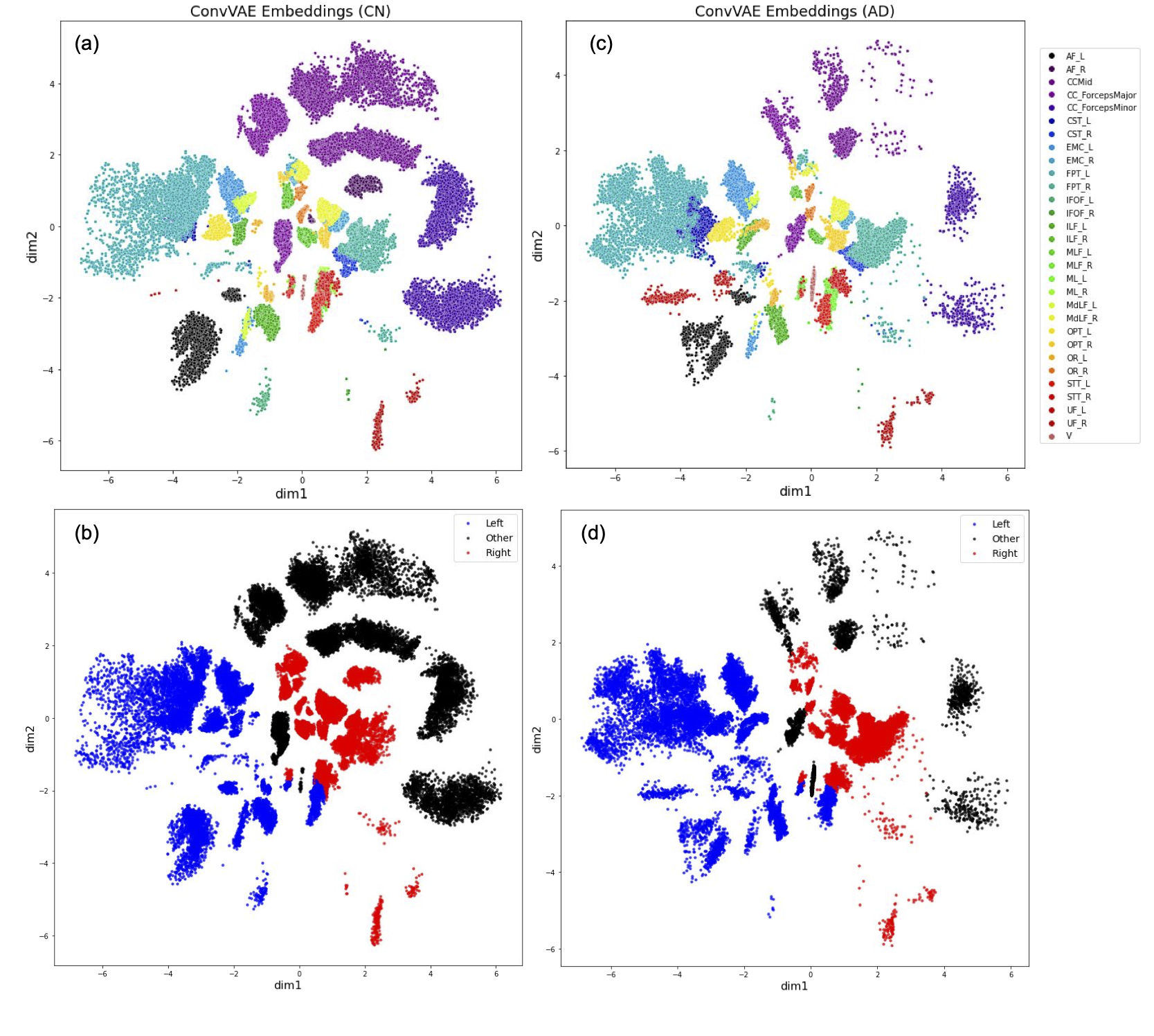
This figure plots 2D streamlines embeddings generated from a ConvVAE trained on a healthy control subject from ADNI3.
- (a)-(b): Embeddings of the CN subject data used to train the ConvVAE model colored by bundles and hemispheres
- (c)-(d): Embeddings of data from a new randomly selected AD subject used in inference colored by bundles and hemispheres.
Figure (a) and (c) show similar cluster patterns for corresponding bundles from two subjects. Figure (b) and (d) shows distinction between left and right hemisphere, with the left hemisphere embeddings spanning more latent space.
This figure shows the 2D embeddings, original streamlines and reconstructed streamlines for 4 select tracts.
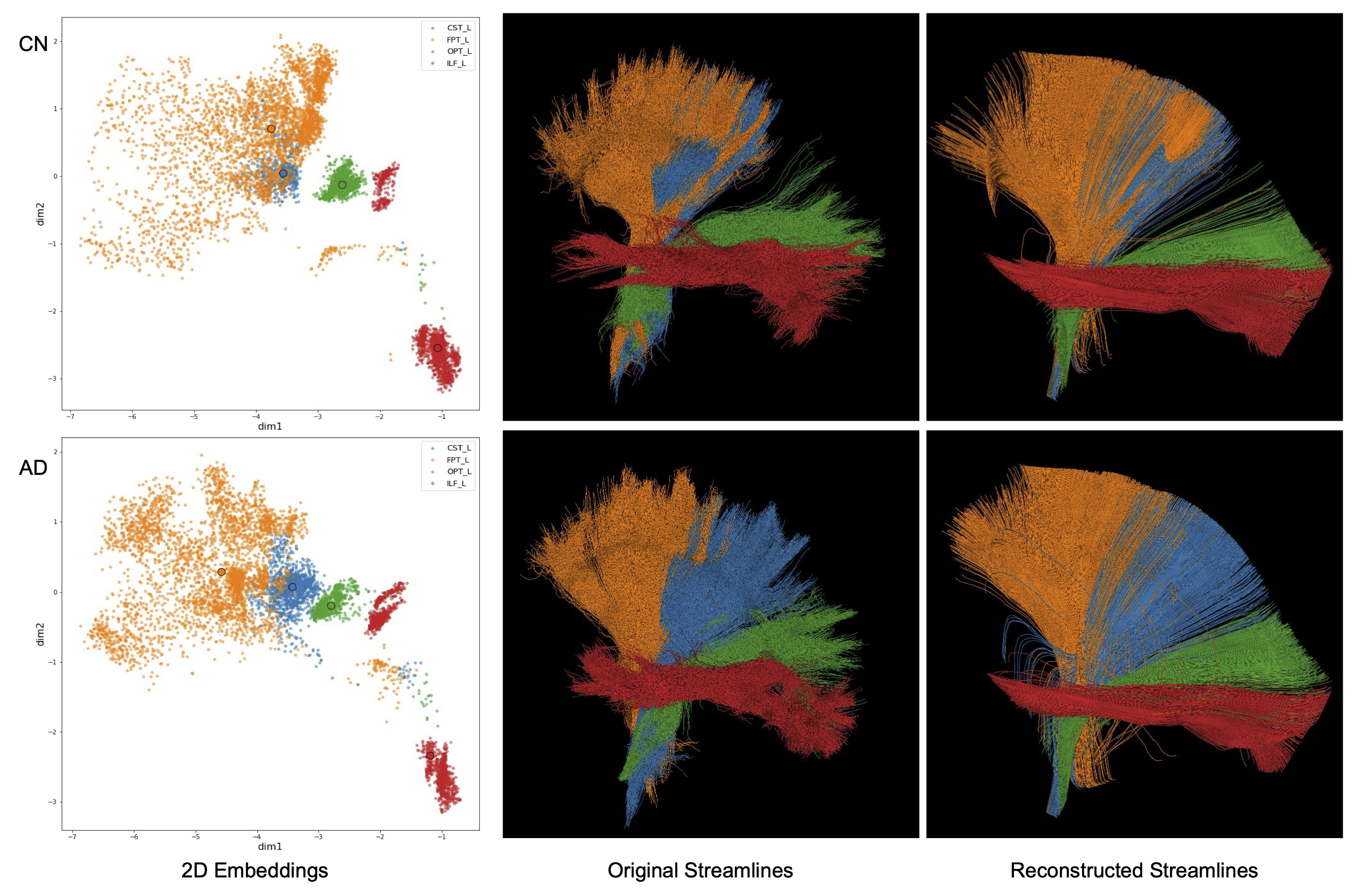
ConvVAE generates 2D embeddings that preserve bundles’ spatial and shape information. It learns a smooth latent space from streamlines, which allows for meaningful decodings from sampled points and can be directly applied to new data. Distance-based algorithms can be used in downstream tasks, such as bundle labeling and intersubject comparisons.
Related Publications
- Feng Y, Chandio BQ, Chattopadhyay T, Thomopoulos SI, Owens-Walton C, Jahanshad N, Garyfallidis E, Thompson PM (2022). Deep generative model for learning tractography streamline embeddings based on Convolutional Variational Autoencoder. ISMRM 2022, London, England, UK, May 07-12, 2022.
Code Availability
- ConvVAE model/training/inference on extracted RecoBundles of single subject (Github).