Deep Multiview Learning for Multimodal Neuroimaging Data
This is a research project I’ve been working on since September 2020 in Dr. Li Shen’s lab at the Perelman School of Medicine, University of Pennsylvania. Our goal is to under Alzheimer’s Disease (AD) structure applying deep multiview learning method on multimodal neuroimaging data.
Motivation
- Multimodal neuroimaging data can reveal complementary information about brain functions, compared to single modality imaging.
- Unsupervised clustering is often used to identify individuals who share similar characteristics, and when applied to neuroimaging or genetics data, can help uncover disease structure or subtypes. However, clustering is often applied to the input features directly, and when working with multimodal data, can suffer from the curse of dimensionality.
Multiview Learning
We propose to use multiview learning methods based on Canonical Correlation Analyis (CCA). Given two input data views \(X_1\in \mathbb{R}^{p_1}\) and \(X_2\in \mathbb{R}^{p_2}\), CCA applies linear transformation \(u_1\in\mathbb{R^{p_1}}\) and \(u_2\in\mathbb{R^{p_2}}\) to each view to maximize correlation between \(u_1^TX_1\) and \(u_2^TX_2\).
To incorporate more than 2 views of data, CCA can be extended to Generalized CCA (GCCA) (Horst, 1961). GCCA learns view-dependent projections \(U_j \in\mathbb{R}^{p_j\times k}\) for each input view \(X_j \in\mathbb{R}^{N\times p_j}\), and view-independent embeddings \(G \in\mathbb{R}^{N\times k}\) that maximizes the canonical correlation, by minimizing \(\Sigma_{j=1}^J \vert\vert G-X_jU_j\vert\vert_2^2\), subjected to \(G^TG=I\).
GCCA can further be extended to Deep Generalized CCA, or DGCCA (Benton, 2017) to apply non-linear transformation, where input views are forward through deep neural networks before applying GCCA (see architecture diagram below).
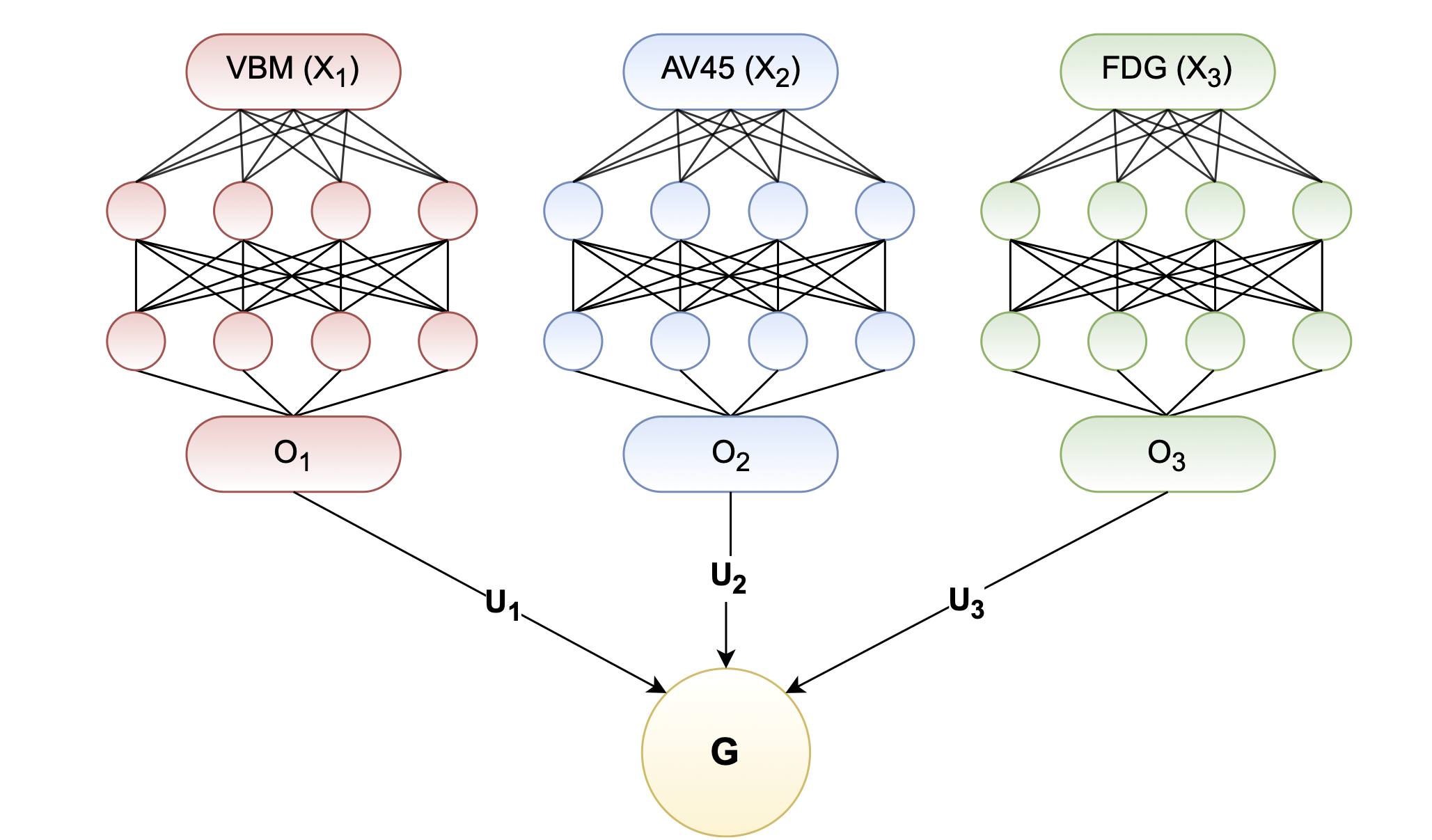
Identifying Population Structure
We apply CCA-based multiview learning on multimodal neuroimaging data collected from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), including sMRI processed using voxel based morphometry (VBM), amyloid-PET (AV45) and FDG-PET, from both cases and controls.
We found that the view-independent embeddings \(G\) learned from DGCCA is shown to effectively capture more variance in the original data than that from generalized CCA (GCCA) which applies only linear transformation. Clusters generated from the DGCCA shared embeddings shows a high level of alignment with the existing case control group. Further genetic association analyses of the clustering results demonstrate that embeddings learned from DGCCA is able to identify promising population structure with a stronger genetic basis.
Publications/Presentations
- Proceedings at the 20th IEEE International Conference on Bioinformatics and Bioengineering (BIBE) in October 2020;
- Poster and presentation at the Penn Department of Biostatistic, Epidemiology & Informatics (DBEI) Research Day in March 2021.
Identifying Disease Subtypes
While multiview learning method DGCCA shows promise in identifying population structure in AD, we further validate our framework to see if it can identify disease subtypes. We applies the same method to Mild Cognitive Impairment (MCI) participants, who are more at risk to develop AD. Existing MCI groups are early MCI (EMCI) and late MCI (LMCI), and we want to see if multimodal imaging data can generate subtypes different from existing groups, and reveal new information.
We generated 2 subtypes from DGCCA embeddings using unsupervised cluster and compared them with the original EMCI and LMCI labels. Our validation studies show that DGCCA subtypes have differential measures in 5 cognitive assessments, 6 brain volume measures and conversion to AD patterns. These subtypes also confirmed AD markers that EMCI and LMCI did not.
Publications/Presentations